Fluorescent Dyes
An Overview

A basic principle in fluorescence microscopy is the highly specific visualization of cellular components with the help of a fluorescent agent. This can be a fluorescent protein – for example GFP – genetically linked to the protein of interest. If cloning is impossible – for instance in histologic samples – techniques such as immunofluorescence staining are used to visualize the protein of interest. For this purpose, antibodies are utilized, which are linked to distinct fluorescent dyes and bind to the adequate target structure either directly or indirectly. With the help of fluorescent dyes, fluorescence microscopy is not only restricted to proteins but can also be used to detect nucleic acids, glycans and other structures. You can even use an application-specific variety of live cell dyes available that allow for organelle visualization with organelle-selective stains (e.g. ER, mitochondria, Golgi) or function assays like, e.g. live cell tracking, labeling, cell proliferation, or live dead assays, where fluorescence is the way of read-out. Even non-biological substances like Calcium ions can be detected. This article provides an introduction to the commonly used fluorescent agents.
Immunofluorescence
In fluorescence microscopy there are two ways to visualize your protein of interest. Either with the help of an intrinsic fluorescent signal - by genetically linking a fluorescent protein to a target protein - or with the help of fluorescently labeled antibodies that bind specifically to a protein of interest. For some biological questions it is more useful or even necessary to perform the latter one. In the case of histological samples, for example, it is not possible to use fluorescent proteins because in general the specimen is derived from an organism which does not hold any fluorescent proteins. Furthermore, if a functioning antibody is available, immunofluorescence is much faster than fluorescent protein techniques, where you have to clone the gene of interest and transfect DNA into the adequate cell. Another disadvantage of fluorescent proteins lies in their nature of being a protein themselves. With it, they have specific proteinous characteristics inside a cell which can lead to dysfunction or misinterpretations concerning the attached protein of interest. However, it should be considered that using fluorescent proteins is generally the method of choice to study living cells.
Immunofluorescence makes use of the very specific binding affinity of an antibody to its antigen. This can have two different appearances. The easiest way is to use one fluorescently labeled antibody which is binding to the protein of interest. This is called direct immunofluorescence.
In most cases there are two forms of antibodies used. The first one binds to the target protein and is not fluorescently labeled itself (primary antibody). But a second one (secondary antibody) which is binding the primary antibody specifically carries a fluorescent dye. This method is called indirect immunofluorescence and has several advantages. On the one hand there is an amplifying effect, because more than one secondary antibody binds to one primary antibody. On the other hand, it is generally easier to find secondary antibodies than the specific primary antibody labeled with a common fluorescent dye. Below, we review the most commonly used fluorescent dyes.
FITC and TRITC
Fluorescein isothiocyanate (FITC) is an organic fluorescent dye and probably one of the most commonly used in immunofluorescence and flow cytometry. It has an excitation/emission peak at 495/517 nm and can be coupled to distinct antibodies with the help of its reactive isothiocyanate group, which is binding to amino, sulfhydryl, imidazoyl, tyrosyl or carbonyl groups on proteins. FITC (was one of the first dyes which was used for fluorescence microscopy and served as a precursor for other fluorescent dyes like Alexa Fluor®488. Its fluorescence activity is due to its large conjugated aromatic electron system, which is excited by light in the blue spectrum.

Fig. 1: Drosophila embryo development, Green: FITC, Red: TRITC.
A dye very often used in combination with FITC is TRITC (Tetramethylrhodamine-5-(and 6)-isothiocyanate). In contrast to FITC, TRITC is not a fluorescein but a derivate of the Rhodamine family. Rhodamines also have a large conjugated aromatic electron system, what leads to their fluorescent behavior. TRITC is excited with light in the green spectrum with a maximum at 550 nm. Its emission maximum is lying at 573 nm. The bond to proteins (e.g. antibodies) is also based on a reactive isothiocyanate group.
Even though FITC and TRITC are still widely used, they are rather weak fluorescent dyes and not recommended for state-of-the-art microscopy.
Cyanines
Alexa Fluor® dyes are a big group of negatively charged and hydrophilic fluorescent dyes, frequently used in fluorescence microscopy. All the Alexa Fluor® dyes are sulfonated forms of different basic fluorescent substances like fluorescein, coumarin, cyanine or rhodamine (e.g. Alexa Fluor®546, Alexa Fluor®633). The respective laser excitation wavelength is mentioned in their labeling. For example, Alexa Fluor®488, one of the most commonly used dyes, has an excitation maximum at 493 nm, which allows excitation with a standard 488 nm laser, and an emission maximum at 519 nm Alexa Fluor®488 is a fluorescein derivate and has similar properties than FITC. However, it shows better stability, brightness and lower pH sensitivity.

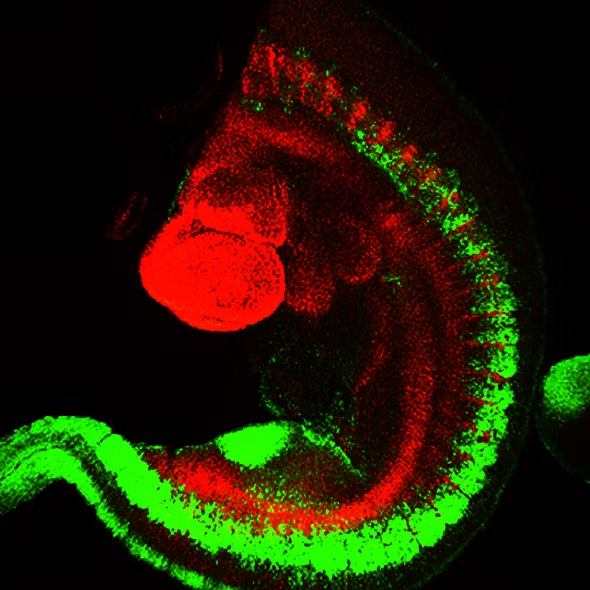
Fig. 2: Mouse transgenic embryo, interlimb somites, Five interlimb somites of an E10.5 mouse transgenic embryo: EpaxialMyf5 eGFP; immuno-stained for GFP-Alexa 488; embryonic muscle fibers stained with Desmin-Cy3, the nuclei are revealed with Hoechst Size from top to bottom: 3.5 mm (a), 800 µm (b). Courtesy of: Aurélie Jory, Cellules Souches et Développement, Institut Pasteur, Paris, France and Imaging centre of IGBMC, IGBMC

Fig. 3: Mouse fibroblasts, Green: F-Actin, FITC, Red: Tubulin, Cy5, Blue: Nuclei, DAPI. Courtesy of: Dr. Günter Giese, Max Planck Institute for Medical Research, Heidelberg, Germany.
DNA staining
You might want to study nucleic acids using fluorescent microscopy. For example, to define the exact position or number of cells by the detection of their nucleus. One of the most common DNA stains is DAPI (4',6-diamidino-2-phenylindole) which binds to A-T rich regions of the DNA double helix. DAPI fluorescence intensity increases if attached to DNA compared to its unbound state. It is excited by UV-light with a maximum at 358 nm. Emission spectrum is broad and peaks at 461 nm. A weak fluorescence can also be detected for RNA binding. In this case, emission shifts to 500 nm. Interestingly, DAPI is able to permeate an intact plasma membrane which makes it useful for fixed and living cells.
A second broadly used DNA stain option is the family of Hoechst dyes. Hoechst 33258, Hoechst 33342, and Hoechst 34580 are Bisbenzimides with intercalating tendency to A-T rich areas. Similar to DAPI they are excited by UV-light and have an emission maximum at 455 nm which is shifted to 510–540 nm in an unbound condition. Hoechst stains are also cell permeable and can therefore be used in fixed and living cells. They exhibit lower toxicity than DAPI.
A membrane-impermeable DNA stain is Propidium-Iodide which is often used to differentiate between living and dead cells in a cell culture because it cannot enter an intact cell. Propidium-Iodide is also an intercalating agent but with no binding preference for distinct bases. In the nucleic acid bound state, its excitation maximum is at 538 nm. Highest emission is at 617 nm. Unbound Propidium-Iodide excitation and emission maxima are shifted to lower wavelengths and lower intensity. It can also bind to RNA without changing its fluorescent characteristics. To distinguish DNA from RNA it is necessary to use adequate nucleases.
A dye which is capable to make a difference between DNA and RNA without previous manipulation is Acridine Orange. Its excitation/emission maximum pair is 502 nm/525 nm in the DNA bound version and turns to 460 nm/650 nm in the RNA bound state. Furthermore, it can enter acidic compartments like lysosomes where the cationic dye is protonated. In this acidic surrounding Acridine Orange is excited by light in the blue spectrum, whereas emission is strongest in the orange region. It is often used to identify apoptotic cells, as they have a lot of engulfed acidic compartments
Compartment and organelle specific dyes
There is a number of specific dyes to study cell compartments such as lysosomes, endosomes or organelles such as mitochondria.
One well known way to observe mitochondria is the utilization of MitoTracker®. This is a cell permeable dye with a mildly thiol-reactive chloromethyl moiety used to bind to matrix proteins covalently by reacting with free thiol groups of cysteine residues. MitoTracker® exists in different colours and modifications . In contrast to conventional mitochondria specific stains like rhodamine 123 or tetramethylrosamine, MitoTracker® is not washed out after destruction of the membrane potential with fixatives.
LysoTracker is a group of dyes available in different colors used to stain acidic compartments such as lysosomes. These are membrane permeable weak bases linked to a fluorophore. Most probably these bases have an affinity to acidic compartments because of protonation.
A comparable compartment to lysosomes is the vacuole in fungi like Saccharomyces cerevisiae. This membrane enclosed space is also of an acidic nature. One way to visualize it in fluorescence microscopy is the use of styryl-based dyes like FM 4-64® or FM 5-95®.
The Endoplasmic Reticulum (ER) is usually stained when studying protein secretion. One classical dye to stain this compartment is DiOC6(3) which has a preference for the ER but still binds to other membranes like those of mitochondria. Another way to specifically stain the ER is to use ER-Trackers like ER-Tracker Green and Red. Both are BODIPY based dyes which are linked to glibenclamide – a sulfonylurease – which binds to ATP sensitive Potassium channels exclusively resident in the ER membrane. BODIPY (boron-dipyrromethene) describes a group of relatively pH insensitive dyes which are almost all water insoluble. This makes them a very good tool for lipid and membrane labeling.

Fig. 4: Purkinje cell, triple labeled parasagittal section of mouse cerebellar cortex. Red: anti-calbindin-D28k/Cy3, Green: anti-GFAP/Cy5, Blue: Hoechst 33258.

Fig. 5: Bovine Pulmonary Endothelial cells. Red: Mitochondria labeled MitoTracker® Red CMXRos, Green: F-actin labeled with green-fluorescent BODIPY® FL phallacidin, Blue: DAPI labeled nuclei. This image was enhanced using 3D blind deconvolution.
The adjacent compartment to the ER – the Golgi apparatus – can be labeled with fluorescent ceramide analogs like NBD C6-ceramide and BODIPY FL C5-ceramide. Ceramides are Sphingolipids which are highly enriched in the Golgi apparatus.
With the help of further lipid-based dyes it is possible to stain special membrane regions like lipid-rafts. These cholesterol rich domains can be visualized by using NBD-6 Cholestrol or NBP-12 Cholesterol amongst others Avanti Polar Lipids.
It is also possible to stain the area of interest with the help of proteins with preferences for distinct locations in the cell. One example is the use of wheat germ agglutinin (WGA) which binds specifically to sialic acid and N-acetylglucosaminyl present in the plasma membrane.
Ion imaging
In the case of neuronal studies, gene activity or cellular movement it is of interest to study the ion concentration of the cell. Sodium, calcium, chloride or magnesium ions have a deep impact on many different cellular events. Typically, ions can be trapped with the help of fluorescently labeled chelators that change their spectral properties when bound to the appropriate ions. One example of labelled chelators are the calcium indicators fura-2, indo-1, fluo-3, fluo-4 and Calcium-Green. For sodium detection, SBFI (sodium-binding benzofurzanisophthalate) or Sodium Green are commonly used. PBFI (potassium-binding benzofurzanisophthalate) detects potassium ions.
Functional assays
“Functional assays” is the broad term used to cover standardized experiments to assess various functions that can be visualized with fluorescent markers. These markers can encompass but is not limited to any of the above-mentioned labeling techniques and fluorophores. For many of these functional assays, staining kits are commercially available and can easily be applied to a vast variety of samples. One example for a functional assay is the commonly known and widely used live dead assay. Two fluorophores are utilized to label live cells and dead cells. Having both values at hand the overall health status of the cells can be assessed. Correlating this information with additional markers may even increase the understanding of the underly process.
Fluorescent dyes and their excitation and emission wavelength peaks
The table below shows a comprehensive list of fluorescent dyes with their respective excitation and emission wavelength peaks. Please note that besides those peaks, every dye features distinct excitation and emission spectra. When selecting several dyes to use in combination in one experiment, researchers should be aware of overlapping excitation and emission spectra due to crosstalk (or bleedthrough), which can result in false negatives or positives, or otherwise obscure data. Another source that can distort fluorescence imaging is autofluorescence by naturally occurring fluorescent proteins in cells and tissues, which particularly needs to be considered in experiments with plants or algae. Good understanding of the spectra of the dyes used in the sample is also important to choose the right light source for excitation (e.g. LED, arc lamps, laser lines) and the right filters and detectors for emission.

Fig. 6: Fluorescence emission profile of Alexa 488 (green curve) and Alexa 555 (yellow curve). The overlap of the two emissions spectra is plain to see. The red line indicates the band pass of a 488 emissions filter.
Table 1
The original article was published by Leica Microsystems.